-Search query
-Search result
Showing all 50 items for (author: chang & cw)

EMDB-36473: 
Cryo-EM structure of the full-length African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide
Method: single particle / : Chang CW, Tsai MD

EMDB-35609: 
Cryo-EM structure of phosphoketolase from Bifidobacterium longum in octameric assembly
Method: single particle / : Chang CW, Tsai MD

EMDB-35610: 
Cryo-EM structure of phosphoketolase from Bifidobacterium longum in dimeric assembly
Method: single particle / : Chang CW, Tsai MD

EMDB-35611: 
Cryo-EM structure of cyanobacteria phosphoketolase complexed with AMPPNP in dimeric assembly
Method: single particle / : Chang CW, Tsai MD

EMDB-35612: 
Cryo-EM structure of cyanobacteria phosphoketolase complexed with AMPPNPin dodecameric assembly
Method: single particle / : Chang CW, Tsai MD

EMDB-35613: 
Cryo-EM structure of cyanobacteria phosphoketolase
Method: single particle / : Chang CW, Tsai MD

EMDB-35617: 
Cryo-EM structure of cyanobacteria phosphoketolase in dodecameric assembly
Method: single particle / : Chang CW, Tsai MD

EMDB-35274: 
The PKR and E3L complex
Method: single particle / : Han CW, Kim HJ

EMDB-33393: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33394: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33392: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33391: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33395: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33396: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33397: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33398: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation)
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-33399: 
Consensus map of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~6.9
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS
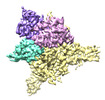
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
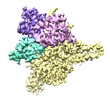
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

EMDB-31495: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ

EMDB-31496: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in nanodiscs with soybean lipids at pH ~8.0
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-31497: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels
Method: single particle / : Lee HJ, Cha HJ, Jeong H, Lee SN, Lee CW, Woo JS

EMDB-32832: 
SARS-CoV-2 Spike in complex with Fab of m31A7
Method: single particle / : Wu YM, Chen X

EMDB-32825: 
Negative stain volume of the mono-GlcNAc-decorated SARS-CoV-2 Spike
Method: single particle / : Chen X, Huang HY

EMDB-30857: 
Human calcium sensing receptor adopts an inactive open-open conformation
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Liu L, Sun DM, Shi CW

EMDB-30858: 
Human calcium sensing receptor adopts an inactive open-closed conformation
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Liu L, Sun DM, Shi CW

EMDB-23589: 
Cryo-EM structure of 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.RnS2 N160K HIV-1 Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-31470: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

EMDB-31471: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

EMDB-30853: 
Human Calcium-Sensing Receptor bound with calcium ions
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Sun DM, Liu L, Shi CW

EMDB-30854: 
Human Calcium-Sensing Receptor bound with L-Trp
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Liu L, Sun DM, Shi CW

EMDB-30855: 
Human Calcium-Sensing Receptor bound with L-Trp and calcium ions
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Liu L, Sun DM, Shi CW

EMDB-30856: 
Human Calcium-Sensing Receptor in the inactive close-close conformation
Method: single particle / : Ling SL, Tian CL, Shi P, Liu SL, Meng XY, Liu L, Sun DM, Shi CW

EMDB-20569: 
Monoclonal antibody 1C01 binds stem domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-20570: 
Monoclonal antibody 1F03 binds head domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-20571: 
Monoclonal antibody 1H09 binds head domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-8977: 
Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Eng ET, Kwong PD

EMDB-20191: 
Cryo-EM structure of vaccine-elicited antibody 0PV-b.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122
Method: single particle / : Gorman J, Kwong PD

EMDB-20189: 
Cryo-EM structure of vaccine-elicited antibody 0PV-a.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122
Method: single particle / : Gorman J, Kwong PD

EMDB-9189: 
Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Xu K, Kwong PD

EMDB-9319: 
Cryo-EM structure at 4.0 A resolution of vaccine-elicited antibody A12V163-a.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Kwong PD

EMDB-9320: 
Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody DFPH-a.15 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Kwong PD

EMDB-9359: 
Cryo-EM structure of vaccine-elicited antibody 0PV-c.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122
Method: single particle / : Gorman J, Kwong PD

EMDB-20017: 
Structure of the rice hyperosmolality-gated ion channel OSCA1.2
Method: single particle / : Maity K, Heumann JM

EMDB-7459: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Carragher B, Potter CS, Kwong PD

EMDB-7460: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Carragher B, Potter CS, Kwong PD

EMDB-8420: 
Cryo-EM structure of BG505 DS-SOSIP HIV-1 Env trimer in complex with vaccine elicited, fusion peptide-directed antibody vFP1.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-8421: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-8422: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-8125: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV fusion peptide targeting N123-VRC34.01 Fab
Method: single particle / : Ozorowski G, Ward AB
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
